Restructuring seed storage proteins for use in food and materials
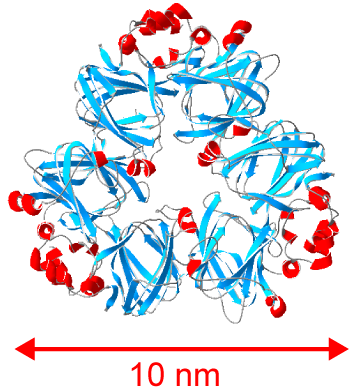
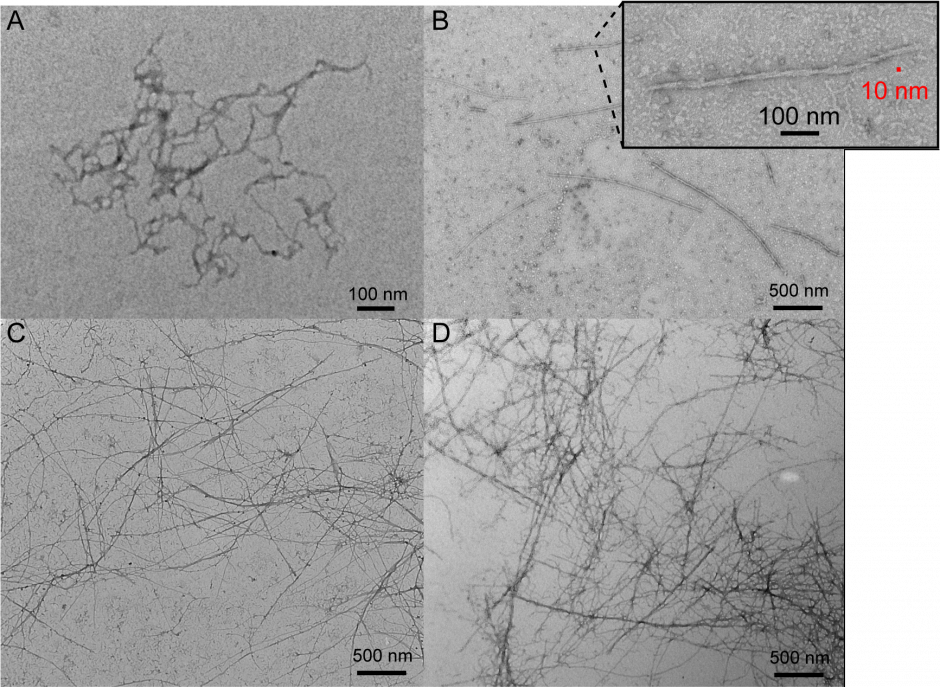
There is growing interest in finding suitable alternatives to animal-derived proteins (e.g. milk, meat and eggs) for use in food. Proteins from legumes (such as soy, pea, lentil and chickpea) could meet this demand if novel methods are found to improve their functionality in food. The functionality (e.g. texture, gelling and foaming ability, and water and fat binding capacity) of plant proteins is generally lower than that of animal protein and can result in foods with lower consumer acceptability. One promising approach to dramatically alter the structure and functionality of proteins is to induce their self-assembly into long, thin strands called ‘nanofibrils’. Nanofibrils are long (μm), thin (10 nm) strands comprised of thousands of protein molecules that undergo dramatic structural changes from their native form and self-assemble together into nanofibrils under the correct conditions. Fibril assembly can be very sensitive to environmental or solution conditions, and the same protein can be induced to form different fibrils or ‘polymorphs’ that have distinct structures, surface chemistries and physical properties. We are examining basic questions around how seed storage proteins assemble into these amyloid-like, nanofibrils, and are defining their structure-function relationships.
Single molecule biophysics
Seeing Beneath the Ensemble Average
Methods that examine samples in bulk (e.g. in solid, crystal or solution form) report on the ensemble average of the molecules. Ensemble averaging occurs over the more than trillions of individual molecules in the sample and can obscure key structural features that are rare or short-lived. By making repeated observations of a single molecule, the entire range of molecular behaviour can be characterized, rather than just the average state. At the single-molecule limit, probability distributions can be measured directly, which are useful to derive biophysical mechanisms, examine molecular heterogeneity, and reveal complex behaviour of biological systems and soft-condensed matter.
We use a dual-trap optical tweezers (the Lumicks C-trap G2) to resolve protein conformational changes at the single-molecule level. A microfluidic system allows us to manipulate single molecules in different solutions.The development of optical tweezers to study biological systems was recognised by the Nobel’s 2018 physics prize.
Optical tweezers can even do tetris!
Assembling a single molecule experiment with optical tweezers:
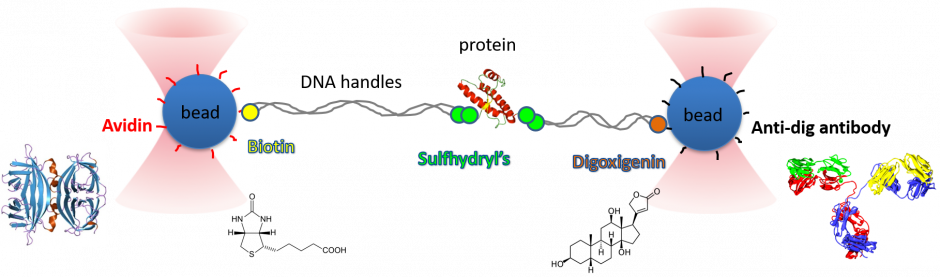
The key to our single molecule experiments is in the molecular construct. We assemble a protein-dsDNA chimera using established chemistries, such as thiol to cysteine attachments. The DNA is then attached to functionalised beads, usually polystyrene or another transparent material with a large index of refraction.
Why dsDNA? It is a relatively stiff polymer that can be made of designed lengths by PCR, while its elastic behaviour under tension has been extensively characterised since the mid-1990s (e.g. Smith 1996).
With the beads trapped by the lasers, we can then stretch the protein out and relax it again, and then repeat the process, measuring the force to unfold/refold the protein with each pull. By doing this many times, we can get a picture of the proteins conformational energy landscape.
Protein Folding Dynamics
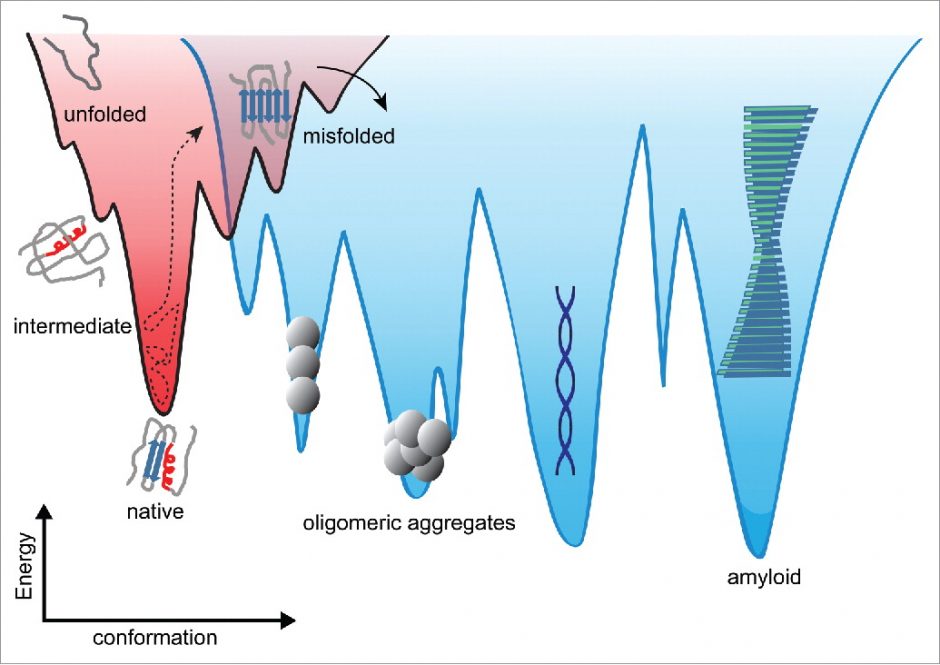
How do proteins fold?
Conformational energy landscapes underly biomolecular behaviour, such as structure, thermodynamic and kinetic stability, binding affinity, dynamics, and functionality. The energetic barrier(s) that separate different conformational states set the time-scales for conformational change, e.g. the stability of an enzyme, or the resistance to misfolding and aggregating into pathogenic amyloid.
Dee, Woodside (2016) Prion, 10:207.
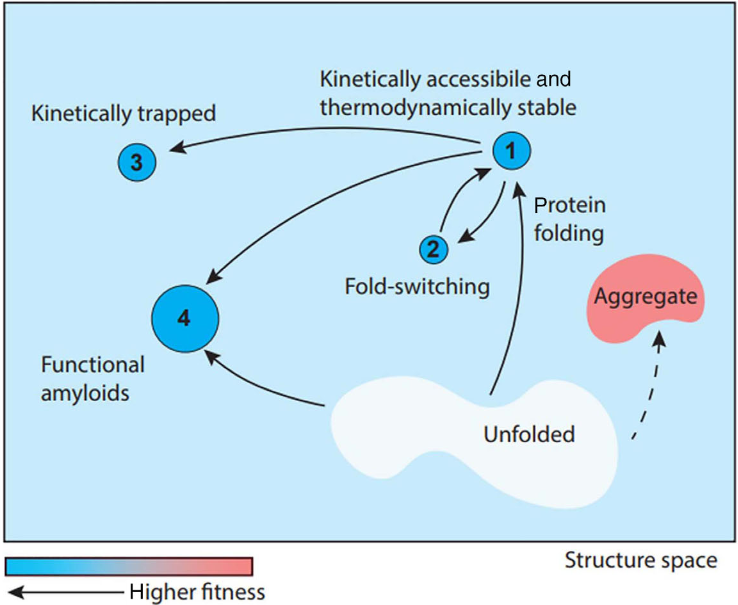
Understanding kinetic barriers and protein stability
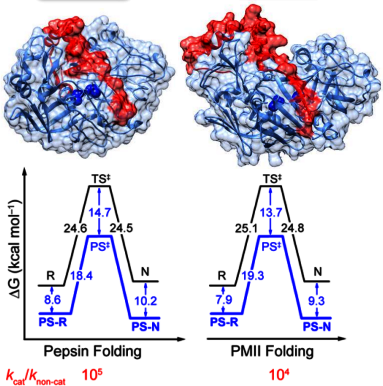
The physical and evolutionary basis of protein stability is poorly understood yet remains critical to many industrial applications using enzymes. The digestive enzyme pepsin and its inactive precursor pepsinogen, which contains an additional prosegment (PS) domain that spans its active site, are an excellent model system to study the nature of folding/unfolding kinetic barriers: pepsin is thermodynamically metastable but kinetically trapped, folding/unfolding on the order of months, whereas pepsinogen is thermodynamically stable and folds/unfolds in minutes. Understanding of how the PS modulates the folding free energy at the single molecule level is expected to provide insights into the microscopic basis of kinetic barriers and the evolution of unfolding resistance. In addition to advancing basic science, a more detailed understanding of protein (un)folding and its evolution has direct benefits for designing industrially-relevant enzymes with increased stability.
Mechanism of amyloid fibril assembly
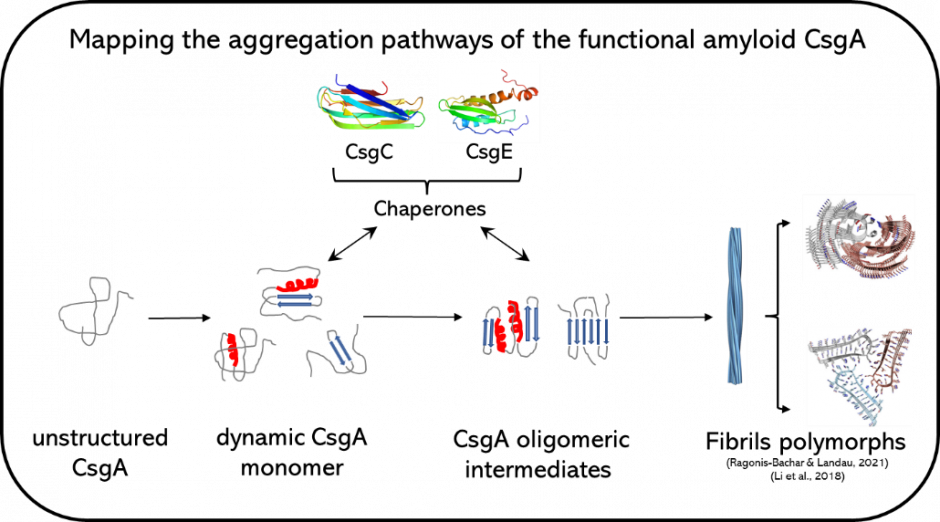
Amyloid fibrils are highly-organized protein aggregates in which the fibril ‘core’ consists of β-sheet structures stacked perpendicular to the long fibril axis. Most proteins are able to convert from their native structure into amyloid forms under certain conditions.
Historically, the best-known amyloids are those associated with diseases, such as Aβ peptide in Alzheimer’s and prion protein in Mad Cow disease (bovine spongiform encephalopathy, vCJD in humans). The role of non-pathogenic, functional amyloids involved in various biological processes in a wide range of species, from bacteria to plants and animals, is becoming evermore apparent. In bacteria, amyloids serve as scaffolds for biofilm assembly, which can protect bacteria from nutritional, mechanical, chemical and physical stresses. Biofilms pose a serious problem to food safety and human health, thus novel drugs targeting amyloid scaffolds are potential anti-biofilm treatments.
Functional Amyloid in Biofilm
Curli amyloid from Escherichia coli involves seven proteins regulating the assembly of CsgA into amyloid fibrils. The molecular details about the CsgA aggregation pathway from monomer to oligomers and to fibril polymorphs remain unknown. CsgA’s high aggregation propensity and intrinsically disordered nature makes structural characterization difficult using bulk techniques owing to ensemble averaging. We are using optical tweezers to resolve the aggregation pathways of CsgA, including the role of various regulating proteins, at the single molecule level. This will provide insights useful for anti-biofilm treatments and for the rational design of functional amyloid used in nanotechnology.